Scientists decipher 11 subtypes in acute leukaemia gene study
The findings could improve clinical trials for testing and developing new AML drugs
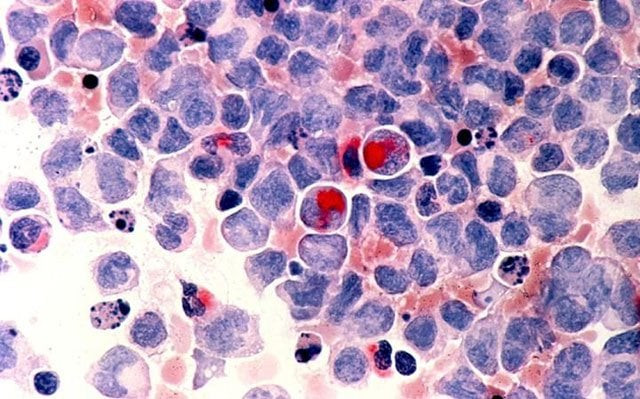
Scientists unpicking the gene faults behind an aggressive blood cancer called acute myeloid leukemia (AML) have found it is not a single disease, but at least 11 different ones with important differences for patients' likely survival chances.
The findings, from the largest study of its kind, could improve clinical trials for testing and developing new AML drugs and change the way patients are diagnosed and treated in future, according to the international team of researchers.
"We have shown that AML is an umbrella term for a group of at least 11 different types of leukemia," said Peter Campbell, who co-led the study from Britain's Wellcome Trust Sanger Institute. "We can now start to decode these genetics to shape clinical trials and develop diagnostics."
From a mutation: Peshawar reports highest number of chronic myeloid leukaemia patients
"What we can clearly see is that a large proportion of what will happen to a patient is written and encoded in the genetic profile of that patient's leukemia," he told reporters at a London briefing.
AML is an aggressive form of blood cancer that develops in cells in the bone marrow and can affect people of all ages. Estimates from the World Health Organization's 2012 GLOBOCAN project suggested some 352,000 people worldwide had AML, and the disease is becoming more prevalent as populations age.
Treatment often involves months of intensive chemotherapy in hospital, but there are large variations in patient survival rates - something doctors and scientists had wanted to investigate.
For this study, published in the New England Journal of Medicine on Wednesday, the team analyzed 1,540 patients with AML. They studied more than 100 genes known to cause leukemia and aimed to seek out common genetic themes behind the development of the disease.
They found the patients could be divided into at least 11 major groups, each with different constellations of genetic changes and distinctive clinical features.
World Chronic Myeloid Leukemia Day: ‘Leukemia common among younger Pakistanis’
Despite identifying common themes, the team also found that most patients had a unique combination of genetic changes driving their leukemia. This genetic complexity helps explain why AML shows such variability in survival rates, they said.
"For the first time we untangled the genetic complexity seen in most AML cancer genomes into distinct evolutionary paths," said Elli Papaemmanuil, who co-led the study at the Memorial Sloan Kettering Cancer Centre in New York.
Mapping Pakistani genetics: Extent of rogue gene’s work in common leukaemia measured
She said having full knowledge of the genetic make-up of a patient's leukemia substantially improved the ability to predict whether that patient could be cured with current drugs.
Such information could also be used to design new trials to develop the best treatments for each AML subtype, she said.


















COMMENTS
Comments are moderated and generally will be posted if they are on-topic and not abusive.
For more information, please see our Comments FAQ